Variable Number of Tandem Repeats
These are segments of DNA that lie next to each other in long strings of repeats. They arise from "slipping" of the DNA polymerase during replication. There are two classes of VNTRs:
Class I
Microsatellites : These are short (3-5bp) segments of DNA that are repeated in long strings of DNA. The polymorphisms arise between individuals within and between populations are are used to look at population structure (within and between) because they are highly variable (polymorphic). Recombinations analyses can also be done to map markers for various dieseases. These markers are extremely helpful because they are under no selection (neutral) which is part of the reason why they are highly variable.
Class II
Minisatellites : These are longer repeats (10-60bp) and are formed in similar ways to microsatelllites and are typically used in DNA fingerprinting. They are used in forensic analysis because they are fail-proof and polymorphic enough to identify individuals.

DNA fingerprinting works on the basis that a certain combination of alleles of minisatellites are specific to each individual and can be used to essentially "barcode" individuals and identify only them. This is useful because it helps forensic scientists identify if an individual was at a crime scene based on their DNA fingerprinting profile.
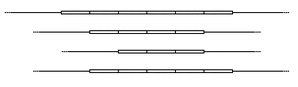
Interpreting DNA fingerprinting data: Each lane on the example gel to the right represents an individual. These could be DNA samples found at the crime scene that you would check against a suspect. Each band represents an allele of a minisatellite. Some individuals will be homozygous for a marker and some will be heterozygous.
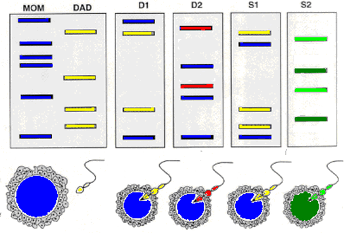
VNTR based fingerprinting also has application in paternity tests as can be seen in the schematic on the right.

Data Interpretation
Data interpretation is done through "fragment analysis". Visualizing the data involves looking at peaks of occurance of the fragment. For example, in the example graph on the right a fragment of size 129 and 135 was identified. If the microsatellite is a 3 bo repeat it would be repeated 45 times in the 135 bp segment and 43 times in the 129 bp segment. These would be considered alleles (and polymorphisms) of this particular microsatellite locus.
Steps of Microsatellite Development
Reference on how microsatellite development is done here
1) Extract DNA
2) Restriction digest into pices
3) Ligate linkers to DNA fragments
4) Dynabead enrighment for microsatellite containing fragments
5) PCR recovery of enriched DNA
6) Ligate enriched DNA into plasmids (and transform)
7) PCR positive colonies, and sequence them