Cre-Lox Technology
Cre-Lox Technology provides a sophisticated means to selectively express a given gene by creating knockouts, conditional knockouts, and reporter strains in a variety of organisms from plants to mice.
How does this technology work?
This technology is contingent upon the presence of two genes Cre and loxP.
Cre Protein: The Cre protein is also known as cyclization recombination protein. It is a 38 kDa protein natively found in the bacterophage P1 as the gene cre. The target substrate for Cre are the loxP regions. Cre will identify pairs of loxP sites and then catalyze reciprocal DNA recombination between the two sites, often resulting in the excision of a small piece of DNA. Cre will only catalyze the recombinantion of two consecutively expressed loxP sites. (In other words, there will not be a loxP site intervening in the middle of the sequence.)
In bacteriophage P1 native Cre has 2 important roles (1) after infection it acts as a backup mechanism for cycling the DNA. (2) Increases the stability of P1 by facilitating separation between dimeric plasmids during bacterial division.
IMPORTANT to note, Cre is NOT the same as CRE (cAMP response element) found in eukaryotes.
LoxP:A loxP site is typically 34bp site. This 34 bp site contains two 13bp inverted repeats with an 8 bp core. The core is a non-pallindromic sequence. All of the 34 bp are not necessary for successful recombination by Cre, some of the base pairs can be modified.
Putting the system together:
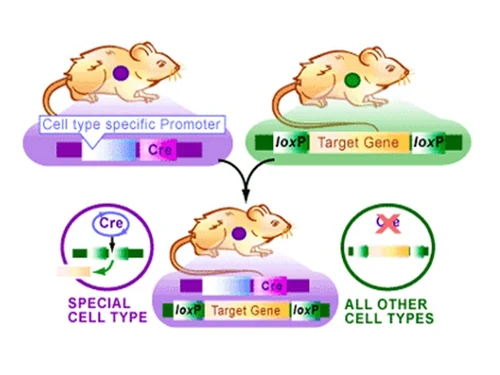
Researchers will select an organism and a target gene they wish to study. Two lox sequences will then be inserted at both ends of the target gene. This DNA will be incoporated into one mouse. The researchers will then decide the conditions under which they want their target gene expressed (all the time vs. part of the time) and based the conditions they will pick another gene and incorporate the Cre gene downstream. This DNA will be incorporated into a second mouse. The two mice will breed and the offspring will have both the cre and loxP sequences. Whenever the promoter preceding the Cre sequence is transcribed, Cre will also be transcribed and later expressed. The expressed Cre will then cleave inbetween both loxP sequences encompassing the target gene and remove the target gene sequence creating a knock-out mouse.
History
The Cre-lox genes were first discovered in P1 bacteriophage in the 1980's. It was discovered that these genes were a part of the virus' normal life cycle and were used to facilitate DNA replication and enable the genomic DNA to circularize. Since then site-specific DNA recombinases have also been observed including in yeast.