No edit summary |
No edit summary |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==='''Cre-Lox Technology'''=== |
==='''Cre-Lox Technology'''=== |
||
| − | Cre-Lox Technology provides a sophisticated means to selectively express a given gene by creating knockouts, conditional knockouts, and reporter strains in a variety of organisms from plants to mice. |
+ | Cre-Lox Technology provides a sophisticated means to selectively express a given gene by creating knockouts, conditional knockouts, and reporter strains in a variety of organisms from plants to mice. |
|
|
||
| Line 61: | Line 61: | ||
<u>'''P1 and the Cre Lox System '''</u> |
<u>'''P1 and the Cre Lox System '''</u> |
||
| − | The bacteriophage DNA is double stranded with a molecular weight of 6.6 x 107, unlike other phages P1 has a linear genetic map. Within the paper, the authors wanted to evaluate the recABC independent recombination on the EcoRI fragment 7. In doing so |
+ | The bacteriophage DNA is double stranded with a molecular weight of 6.6 x 107, unlike other phages P1 has a linear genetic map. Within the paper, the authors wanted to evaluate the recABC independent recombination on the EcoRI fragment 7. In doing so, they uncovered the Cre enzyme and the loxP regions. |
| − | <u>Experimental Setup:</u> Researchers initially were interested in the fact that the |
+ | <u>Experimental Setup:</u> Researchers initially were interested in the fact that the bacteriophage P1 contained a linear genetic map. As other phages were known to have circular genetic maps, they hypothesized that large segments at the ends of P1 were missing genetic markers or that P1 contained a highly favorable genetic recombination region. After mapping the DNA they discovered markers in both ends of the P1 gene map, ruling out their first idea. To assess the second idea, whether P1 had regions that were highly favorable for genetic recombination, they looked at various fragments of the P1 DNA. These fragments were generated using the restriction enzyme EcoRI. Each fragment was given a number and identified as such throughout the remainder of the experiment. |
| + | [[File:Pl_maps.jpg|thumb|left|500px|P1 and Lambda Restriction Maps. "Bacteriophage P1 Site Specific Recombination"]]As seen on the left. The P1 map is at the top and fragment 7 is located at the ends. These fragments would later be incorporated into a lambda vector. The lambda map is at the bottom of the figure to the left. White arrows represent EcoRI restriction sites, while black arrows represent BamHI restriction sites. The specific lambda vector used was actually lambdaDam15 b538 srIh3 cI857(ts) nin5 to which there was only 1 EcoRI restriction site. |
||
| + | After treating P1 with EcoRI, a variety of P1 fragments including fragment 7 and fragment 3 were individually inserted into the single restriction site of the lambdaDam15 b538 srIh3 cI857(ts) nin5. Hybrids were designated h(lambda)-P1:7, h-P1:3, with 7 and 3 signifying the incorportation of the respective P1 fragment. |
||
| + | The hybrid phages were then subjected to in vivo genetic crosses. To do this, bacterial cells were grown and then infected with 3 x 10^8 of each parent phage. After a period of time cells were lysed and phage DNA was analyzed for recombination. Recombination was determined the arrangement or rearrangement of markers that flank a given region. |
||
| + | The same genetic cross was repeated using the hybrids, except lambda and host recombination systems were inactivated by mutation. If recombination were to occur it would need to use features intrinsic to the P1 fragment. Results showed that there was a high frequency of recombination when both parent lambdas contained P1 fragment 7. This lead them to believe that P1 fragment 7 has sites that must be present in both parents for recA-independent recombination to occur. They designated these sites loxP. [[File:Recomb_table.jpg|thumb|left|300px|Recombination Table. "Bacteriophage P1 Site Specific Recombination"]] |
||
| + | |||
| + | |||
| + | The table to the left indicates the recombination observed (right column) using various parent P1 fragments (middle column) in different bacterial strains (left column). Bacterial strains with parent P7 fragments had notably higher levels of recombination. |
||
| + | |||
| + | |||
| + | |||
| + | Upon further investigation, they also found that markers which were not flanking fragment 7 were recombined following a recombination event. |
||
| + | |||
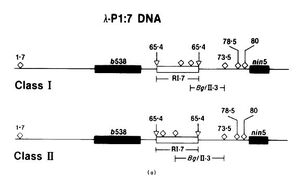
| + | Additionally, the orientation of the fragment 7 insert was also [[File:Class_i_class_ii.jpg|thumb|left|300px|Class I and Class II BglIII arrangements. "Bacteriophage P1 Site-Specific Recombination"]]evaluated. The fragment contains 2 assymetrically located BglII restriction sites. After the fragment has been digested with BglIII, 2 possible fragments can be generated one designated Class I and the other designated Class II. (See image labeled h-P1:7 DNA). Different classes were observed in the hybrids. When they looked at the recombination frequencies of the different classes they found when both parent hybrids were class II recombination was the highest (14%), followed by parents who both were class I (12%). When the parents had mixed classes, recombination was minimal (0.06-0.04%). |
||
| + | |||
| + | [[File:Creloxp_map.jpg|thumb|left|400px|Cre-LoxP genes in the P1 fragment. "Bacteriophage P1 Site-Specific Recombination"]]To determine the specific location of loxP within fragment 7, deletion mutations were performed. The deletions were isolated and digested with restriction enzymes. The products were run on agarose gels to determine the boundaries of the loxP sites. |
||
| + | |||
| + | Additional mutatants were generated and demonstrated that fragment 7 contained a recombinase gene, that could act on two loxP sites. This recombinase was then called Cre. |
||
| + | |||
| + | The figure (to the left/above) depicts the general region where the cre and loxP genes are found in fragment 7. The black bars below are some of the general deletion mutations generated while trying to determine the location of these elements. |
||
| Line 85: | Line 104: | ||
'''<u>Overview of the Cre-lox system first used in Mammalian Cells</u>''' |
'''<u>Overview of the Cre-lox system first used in Mammalian Cells</u>''' |
||
| − | Researchers, Dr. Brian Sauer and Dr. Nancy Henderson, successfully applied the Cre-Lox system in mouse cell line (C-127) . The cre gene was downstream a MT-I (or mouse metallothionein gene) and upstream from BPV(bovine papilloma virus) a sequence that would enable extrachromosomal replication of the plasmid in the mouse cell. The loxP sites encompassed kan r gene in a plasmid pRH43 (Kan r stands for Kanamycin resistant). |
+ | Researchers, Dr. Brian Sauer and Dr. Nancy Henderson, successfully applied the Cre-Lox system in mouse cell line (C-127) . The cre gene was downstream a MT-I (or mouse metallothionein gene) and upstream from BPV(bovine papilloma virus) a sequence that would enable extrachromosomal replication of the plasmid in the mouse cell. The loxP sites encompassed kan r gene in a plasmid pRH43 (Kan r stands for Kanamycin resistant). |
|
|
||
| Line 92: | Line 111: | ||
===<u>Fields that have used the Cre-lox System</u>=== |
===<u>Fields that have used the Cre-lox System</u>=== |
||
| − | -This use of this system in mice is widespread enabling scientists to study everything from the inner ear to cancers. |
+ | -This use of this system in mice is widespread enabling scientists to study everything from the inner ear to cancers. |
|
|
||
Latest revision as of 06:20, 13 December 2012
Cre-Lox Technology[]
Cre-Lox Technology provides a sophisticated means to selectively express a given gene by creating knockouts, conditional knockouts, and reporter strains in a variety of organisms from plants to mice.
How does this technology work?[]
This technology is contingent upon the presence of two genes Cre and loxP.
Cre Protein: The Cre protein is also known as cyclization recombination protein. It is a 38 kDa protein natively found in the bacterophage P1 as the gene cre. The target substrate for Cre are the loxP regions. Cre will identify pairs of loxP sites and then catalyze reciprocal DNA recombination between the two sites, often resulting in the excision of a small piece of DNA. Cre will only catalyze the recombinantion of two consecutively expressed loxP sites. (In other words, there will not be a loxP site intervening in the middle of the sequence.)
In bacteriophage P1 native Cre has 2 important roles (1) after infection it acts as a backup mechanism for cycling the DNA. (2) Increases the stability of P1 by facilitating separation between dimeric plasmids during bacterial division.
Important to note, Cre is NOT the same as CRE (cAMP response element) found in eukaryotes.
LoxP: A loxP site is typically 34bp site. This 34 bp site

A LoxP site with inverted repeats and core.http://www.springerimages.com/Images/RSS/1-10.1007_978-1-60761-974-1_12-1
contains two 13bp inverted repeats with an 8 bp core. The core is a non-pallindromic sequence. All of the 34 bp are not necessary for successful recombination by Cre, some of the base pairs can be modified.
Putting the system together:[]
Researchers will select an organism and a target gene they wish to study. Two lox
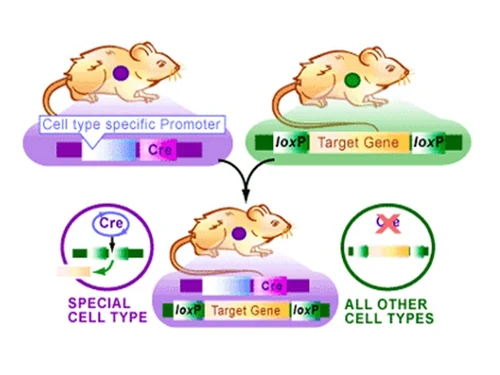
The Cre Lox System in Mice. From the Science Creative Quarterly, Article: TARGETING YOUR DNA WITH THE CRE/LOX SYSTEM By Alfred Pechisker (Aug 2004) http://www.scq.ubc.ca/targeting-your-dna-with-the-crelox-system/
sequences will then be inserted at both ends of the target gene. This DNA will be incoporated into one mouse. The researchers will then decide the conditions under which they want their target gene expressed (never expressed or part of the time expressed) . If they want there target gene to never be expressed theyall theand based the conditions they will pick a gene that is always expressed (like a housekeeping gene)and incorporate the Cre gene downstream. If they only want their target gene to sometimes be expressed they will select a gene that is also sometimes expressed and incorporate the Cre gene downstream. This DNA will be incorporated into a second mouse. The two mice will breed and the offspring will have both the cre and loxP sequences. Whenever the promoter preceding the Cre sequence is transcribed, Cre will also be transcribed and later expressed. The expressed Cre will then cleave inbetween both loxP sequences encompassing the target gene and remove the target gene sequence. So if one opted to place the cre gene downstream of a housekeeping gene, the housekeeping gene and cre would always be expressed. If Cre was always expressed, the target gene would always be excised so no expression of the target gene would occur.
Generation of Different Mutations[]
Depending on the
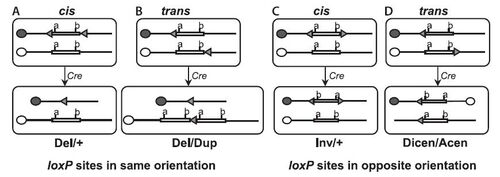
Mutations due to varied orientation of the loxp sites. Cre/loxP-Mediated Chromosome Engineering of the Mouse Genome.V. Brault · V. Besson · L. Magnol · A.Duchon · Y. Hérault
orientation of the loxp sites different mutations can be generated. As you can see to the left Deletions, Duplications, and Inversions are some of the mutations researchers can generate.
History[]
The Cre-lox genes were first discovered in P1 bacteriophage in the 1980's. It was discovered that these genes were a part of the virus' normal life cycle and were used to facilitate DNA replication and enable the genomic DNA to circularize. Since then site-specific DNA recombinases have also been observed including in yeast.
Historic Papers of Note -Bacteriophage Pl Site-specific Recombination I. Recombination between loxP Sites. Nat Sternberg, Daniel Hamilton. (1981) - Discovery of the Cre-lox system in bacteriophage P1.
- Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae.- Sauer B. (1987) - first use of Cre recombinase in yeast.
- Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1- Sauer B, Henderson N (1988) - first use of Cre recombinase in mammalian cells.
-Tissue-and site-specific DNA recombination in transgenic mice - Paul Orban, Daniel Chui, Jamey Marth (1992) - first use of Cre-loxP mouse.
Brief Overview of the Early Papers[]
P1 and the Cre Lox System
The bacteriophage DNA is double stranded with a molecular weight of 6.6 x 107, unlike other phages P1 has a linear genetic map. Within the paper, the authors wanted to evaluate the recABC independent recombination on the EcoRI fragment 7. In doing so, they uncovered the Cre enzyme and the loxP regions.
Experimental Setup: Researchers initially were interested in the fact that the bacteriophage P1 contained a linear genetic map. As other phages were known to have circular genetic maps, they hypothesized that large segments at the ends of P1 were missing genetic markers or that P1 contained a highly favorable genetic recombination region. After mapping the DNA they discovered markers in both ends of the P1 gene map, ruling out their first idea. To assess the second idea, whether P1 had regions that were highly favorable for genetic recombination, they looked at various fragments of the P1 DNA. These fragments were generated using the restriction enzyme EcoRI. Each fragment was given a number and identified as such throughout the remainder of the experiment.
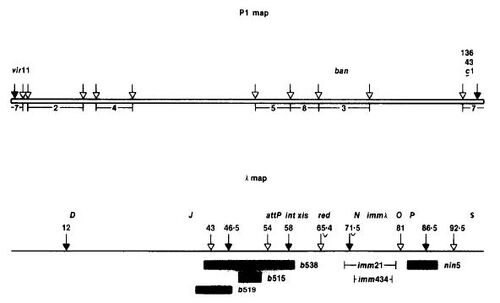
P1 and Lambda Restriction Maps. "Bacteriophage P1 Site Specific Recombination"
As seen on the left. The P1 map is at the top and fragment 7 is located at the ends. These fragments would later be incorporated into a lambda vector. The lambda map is at the bottom of the figure to the left. White arrows represent EcoRI restriction sites, while black arrows represent BamHI restriction sites. The specific lambda vector used was actually lambdaDam15 b538 srIh3 cI857(ts) nin5 to which there was only 1 EcoRI restriction site.
After treating P1 with EcoRI, a variety of P1 fragments including fragment 7 and fragment 3 were individually inserted into the single restriction site of the lambdaDam15 b538 srIh3 cI857(ts) nin5. Hybrids were designated h(lambda)-P1:7, h-P1:3, with 7 and 3 signifying the incorportation of the respective P1 fragment.
The hybrid phages were then subjected to in vivo genetic crosses. To do this, bacterial cells were grown and then infected with 3 x 10^8 of each parent phage. After a period of time cells were lysed and phage DNA was analyzed for recombination. Recombination was determined the arrangement or rearrangement of markers that flank a given region.
The same genetic cross was repeated using the hybrids, except lambda and host recombination systems were inactivated by mutation. If recombination were to occur it would need to use features intrinsic to the P1 fragment. Results showed that there was a high frequency of recombination when both parent lambdas contained P1 fragment 7. This lead them to believe that P1 fragment 7 has sites that must be present in both parents for recA-independent recombination to occur. They designated these sites loxP.
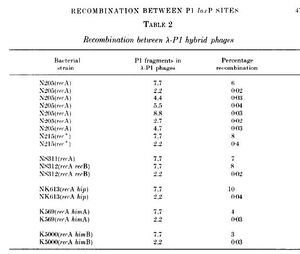
Recombination Table. "Bacteriophage P1 Site Specific Recombination"
The table to the left indicates the recombination observed (right column) using various parent P1 fragments (middle column) in different bacterial strains (left column). Bacterial strains with parent P7 fragments had notably higher levels of recombination.
Upon further investigation, they also found that markers which were not flanking fragment 7 were recombined following a recombination event.
Additionally, the orientation of the fragment 7 insert was also
Class I and Class II BglIII arrangements. "Bacteriophage P1 Site-Specific Recombination"
evaluated. The fragment contains 2 assymetrically located BglII restriction sites. After the fragment has been digested with BglIII, 2 possible fragments can be generated one designated Class I and the other designated Class II. (See image labeled h-P1:7 DNA). Different classes were observed in the hybrids. When they looked at the recombination frequencies of the different classes they found when both parent hybrids were class II recombination was the highest (14%), followed by parents who both were class I (12%). When the parents had mixed classes, recombination was minimal (0.06-0.04%).
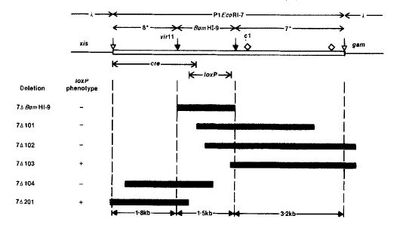
Cre-LoxP genes in the P1 fragment. "Bacteriophage P1 Site-Specific Recombination"
To determine the specific location of loxP within fragment 7, deletion mutations were performed. The deletions were isolated and digested with restriction enzymes. The products were run on agarose gels to determine the boundaries of the loxP sites.
Additional mutatants were generated and demonstrated that fragment 7 contained a recombinase gene, that could act on two loxP sites. This recombinase was then called Cre.
The figure (to the left/above) depicts the general region where the cre and loxP genes are found in fragment 7. The black bars below are some of the general deletion mutations generated while trying to determine the location of these elements.
Overview of Cre-lox first used in Yeast
Researcher Brian Sauer had noted in some instances that bacterial DNA enzymes retained the ability to act on eukaryotic DNA, despite a variety of significant differences between the two types of cells. Having acknowledged this, Dr. Sauer decided to investigate whether a bacteria recombination system could be used in the eukaryotic cell. He selected the cre-lox site specific recombination system found in coliphage P1 (bacteriophage that infects E.coli). This paper was the first paper where researchers began to see the potential to use the cre-lox system in other eukaryotes for site specific recombination.
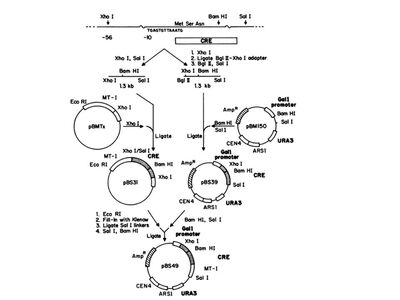
Generating plasmids expressing the Cre. (Functional Expression of the cre-lox Site-Specific Recombination System in the Yeast Saccharomyces cerevisiae. Brian Sauer (1987), Figure 1)
What they actually did in the paper:
Figure to the left demonstrates the construction of the cre gene downstream the yeast galactose gene GALI. To accomplish this they obtained pBS7, which contains the cre gene. From this they were able perform restriction digests to pBS7 to exise the cre gene. They also digested the pBM150 (right side of the figue), which contained the native GALI gene and were able to ligate the two together. They also incorporated cre upstream of a mouse metallothionein gene (the left side of the figure) . The resulting plasmid, pBS49, contained the GALI gene upstream cre followed by the mouse metallothionein gene.
Restriction Digests and ligations were also used to incorporate the loxP sites around the LEU2 gene.
Overview of the Cre-lox system first used in Mammalian Cells
Researchers, Dr. Brian Sauer and Dr. Nancy Henderson, successfully applied the Cre-Lox system in mouse cell line (C-127) . The cre gene was downstream a MT-I (or mouse metallothionein gene) and upstream from BPV(bovine papilloma virus) a sequence that would enable extrachromosomal replication of the plasmid in the mouse cell. The loxP sites encompassed kan r gene in a plasmid pRH43 (Kan r stands for Kanamycin resistant).
Fields that have used the Cre-lox System[]
-This use of this system in mice is widespread enabling scientists to study everything from the inner ear to cancers.
Works Cited:[]
Review Article: Inducible Gene Targeting in Mice Using the Cre/lox System. Brian Sauer (1998)
Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae.- Sauer B. (1987) - first use of Cre recombinase in yeast.
Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1- Sauer B, Henderson N (1988)
Bacteriophage Pl Site-specific Recombination I. Recombination between loxP Sites. Nat Sternberg, Daniel Hamilton. (1981) - Discovery of the Cre-lox system in bacteriophage P1.
Cre/loxP- Mediated Chromosome Engineering of the Mouse Genome. V. Brault, V. Besson, L. Magnol. (2007)